http://genetic.ulriccollette.com/

Similarities
Explanation:
In much the same way that family members resemble each other like in the above photos, if species are related, as evolution purports, one would expect to see similarities between related species. These similarities, especially if they are of traits that don’t seem to give an advantage to a species, allow us to infer a common ancestor that most likely shared this same trait.
Prediction: If species are related, they should share similarities. These similarities should become more frequent and intensify the more closely related species are.
Falsification: Species are completely unique, distinct from each other.
Observations:
There are four main categories of similarities that we will look at:
- Anatomical similarities
- Genetic similarities
- Embryological development similarities
- Geographic distribution similarities
Corroboration: Similarities from multiple disciplines agree. The similarities are non-adaptive, The number and intensity of the similarities increases the more recently the species shared a common ancestor.
Inference: The inference that explains the patterns from the above four categories is that they had a common ancestor that they inherited those traits from.
What is also so important about these similarities listed below is that they aren’t in isolation. All these similarities are nested or hierarchical with many other traits.

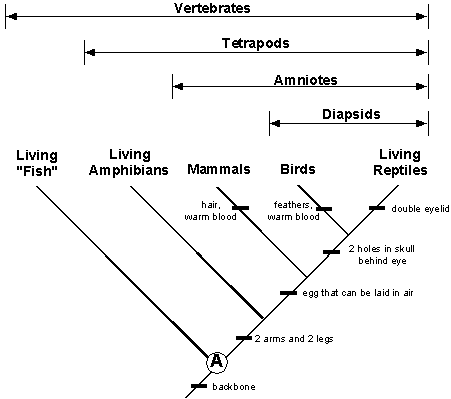
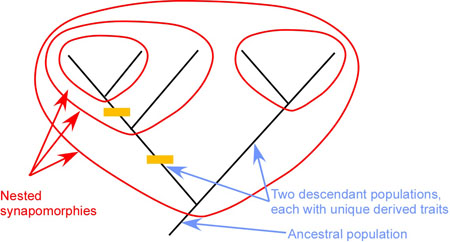
Examples of evidence:
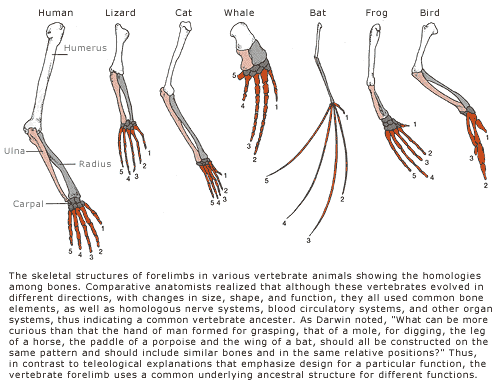
Anatomical: structural similarities between organisms’ body parts.
- Examples: homologous structures, fossils, embryological development, synapomorphy, etc
Orchid homology: Orchids come in a stunning variety of shapes..
Though diverse, these orchids have some major anatomical similarities in common. Each shares the below features:
Tetrapod pentadactyly: The vast majority of vertebrates with four appendages (tetrapod) have five fingers (pentadactyly).

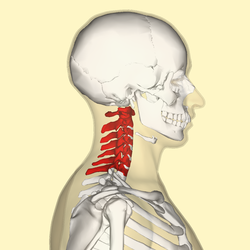
Almost all mammals have 7 neck vertebrae (manatees and sloths are odd exceptions to this–read more here, here, here). While nature likes breaking rules occasionally that still leaves roughly 5,400 species of mammal to back up this general rule. Check out some examples below.

Most insects have 6 legs.

Spider have 8 legs.
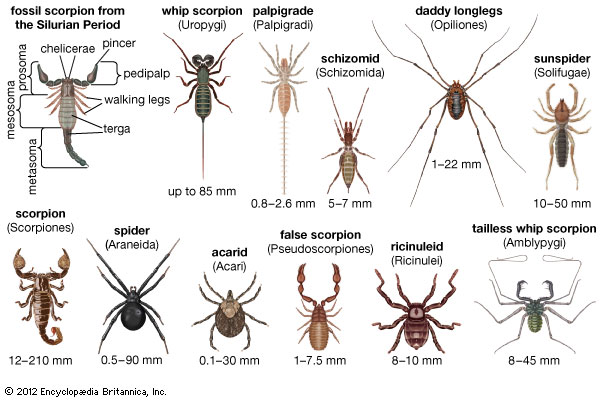
http://kids.britannica.com/comptons/art-178265/Arachnids-are-common-in-different-shapes-and-sizes
Echinoderm pentaradial symmetry: Though this clade is diverse–sea star, sea urchin, sea cucumber, brittle star–they have something in common–they almost all have five sides as adults.

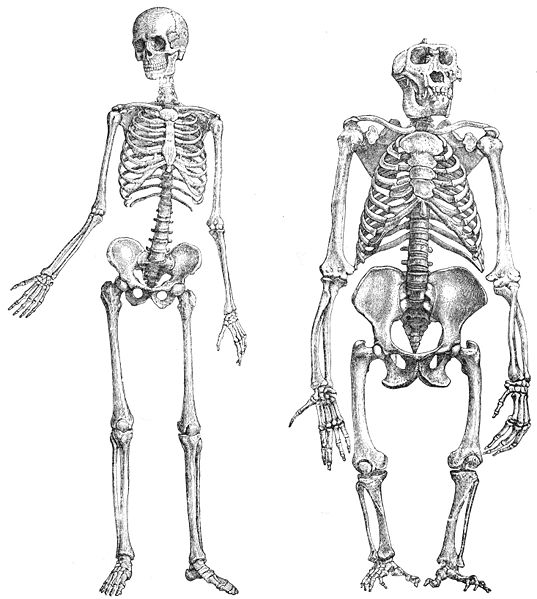
Chimpanzees and humans have nearly identical skeletal structures; they are just stretched in different ways. Explore this here: http://www.pbs.org/wgbh/nova/evolution/compare-skeletons.html
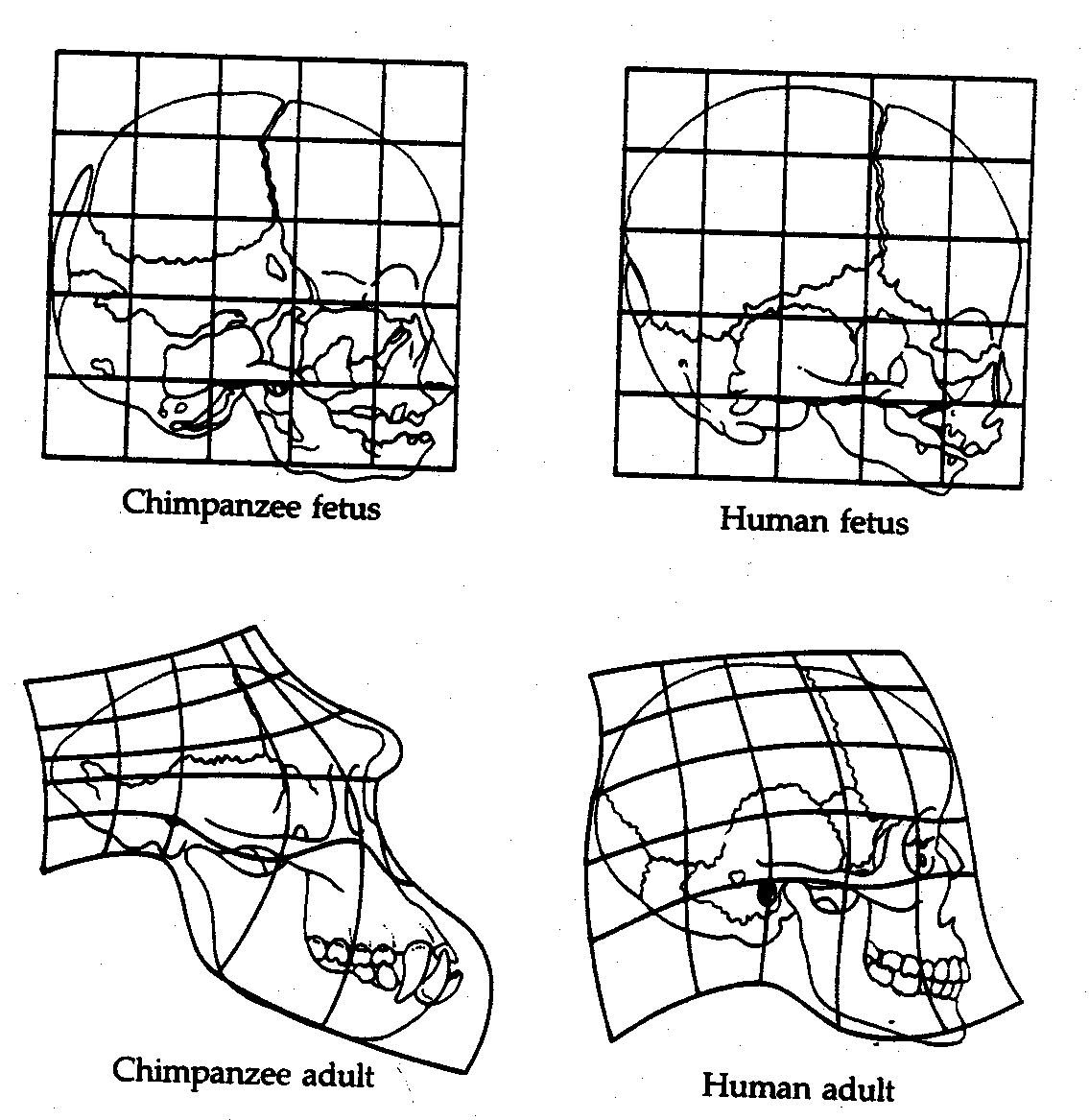
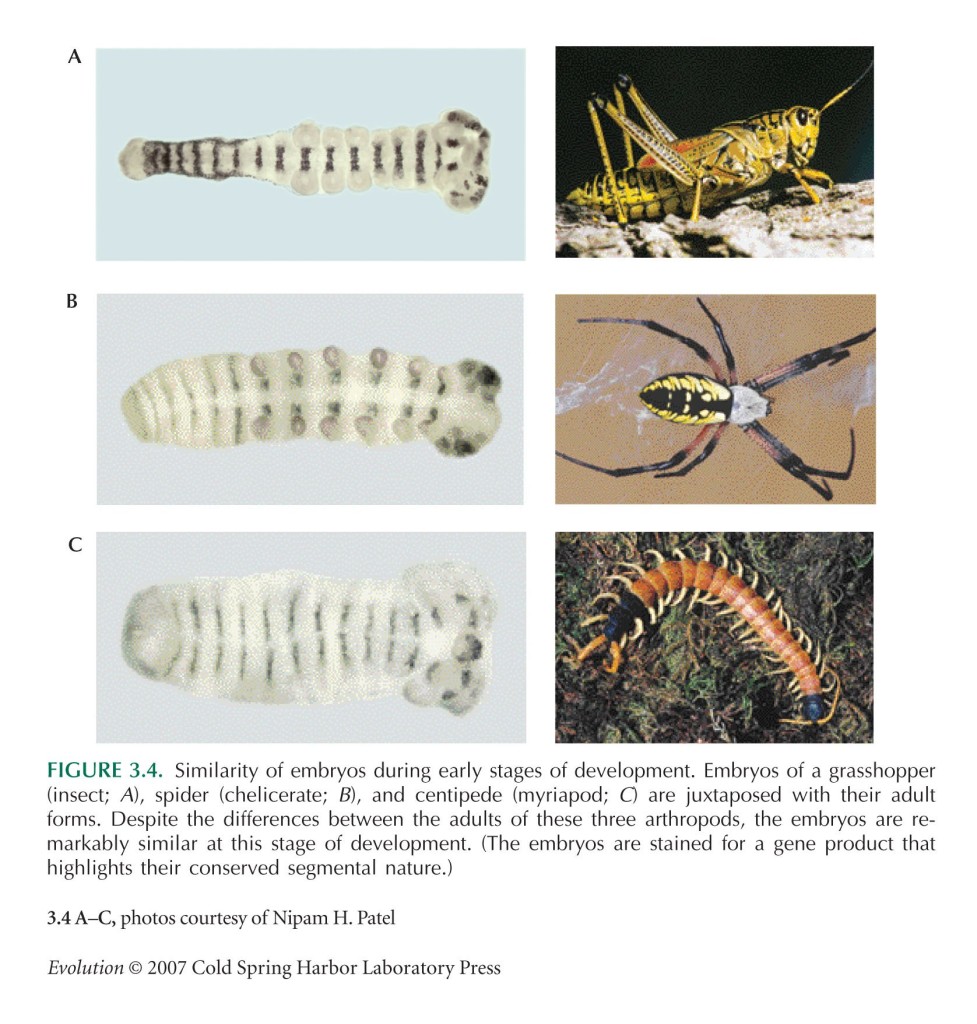
Embryological development similarities (much more on this under ‘remnants‘).

Genetic similarities

Ben Fry
An image of how the gene FOXP2 differs in humans versus chimps. The gene is believed to be one of the primary differences between our species, as it has been linked to language acquisition. All ~75,000 letters of the gene are shown, however only nine letters (shown with red dots) comprise the only significant (coding) changes in the gene. http://benfry.com/humansvschimps/

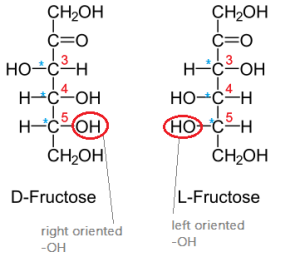
Why should it be that biological sugars are in the ‘D’ orientation? Why not a mix of ‘L’ and ‘D’? Evolution! Our common ancestor had enzymes that operated on ‘D’ and therefore all its descendants (like us!) also uses the same type of sugar.
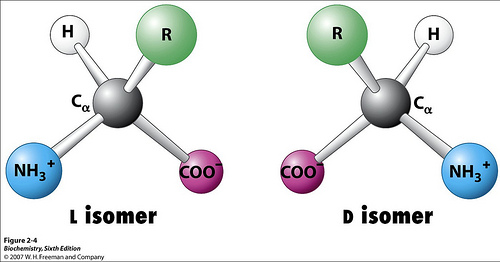
Proteins used in Earth life are almost always ‘L’ conformation (the ‘L’ vs ‘D’ has to do with chirality and the direction that the constituents rotate according to their chemical priority designations when hydrogen is facing away from the observer).
Amino acid or protein sequence similarities: This is a different way of representing genetic similarity (since genes obviously are the precursors to protein sequences). Many genetic mutations in genes do not get represented in protein sequences (since changes in the third codon site are usually synonymous mutations do not result in the translation of a different amino acid).
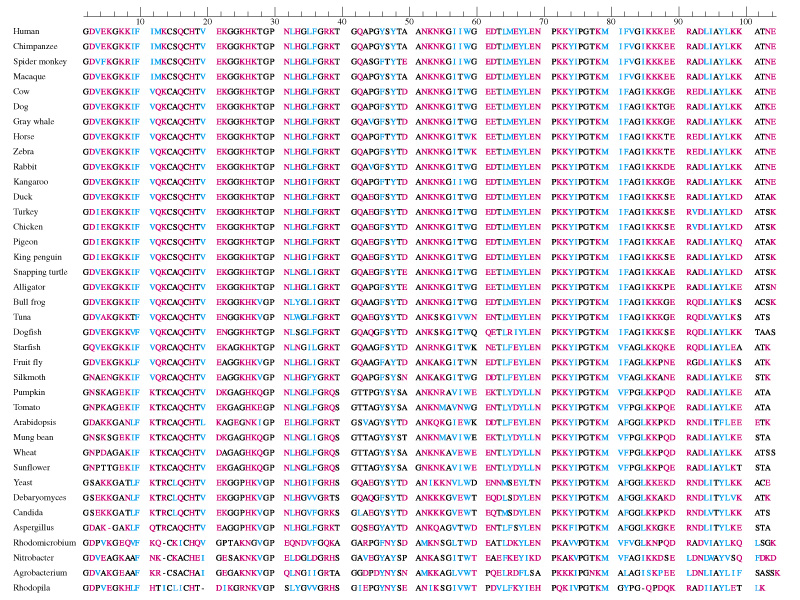

http://biologicalphysics.iop.org/cws/article/lectures/47042
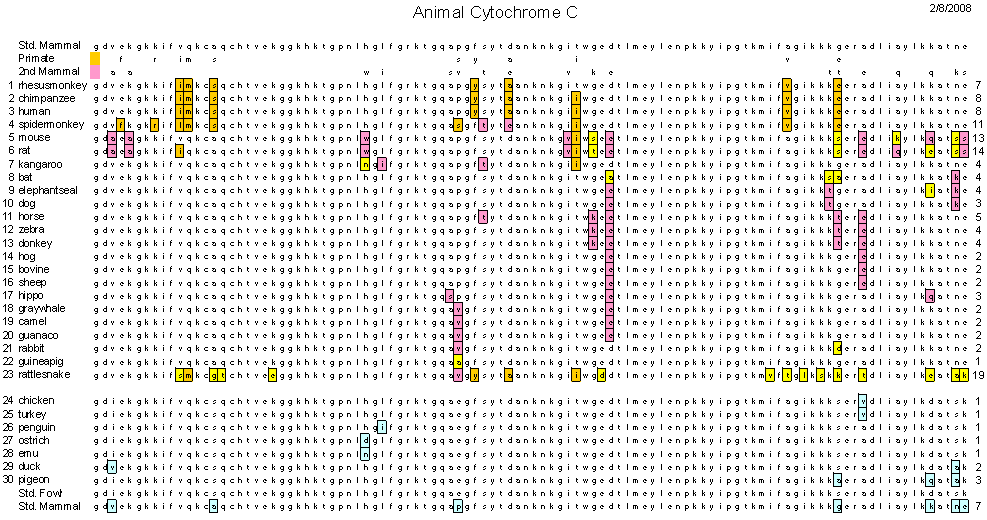
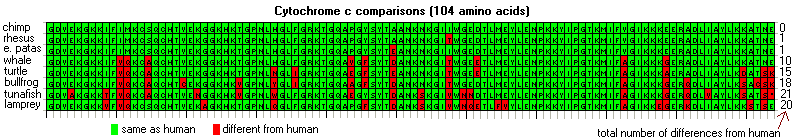
These differences in genetic or protein sequences can easily be used to determine divergence. They are in essence molecular clocks that can be used to ascertain and predict how long ago species diverged.
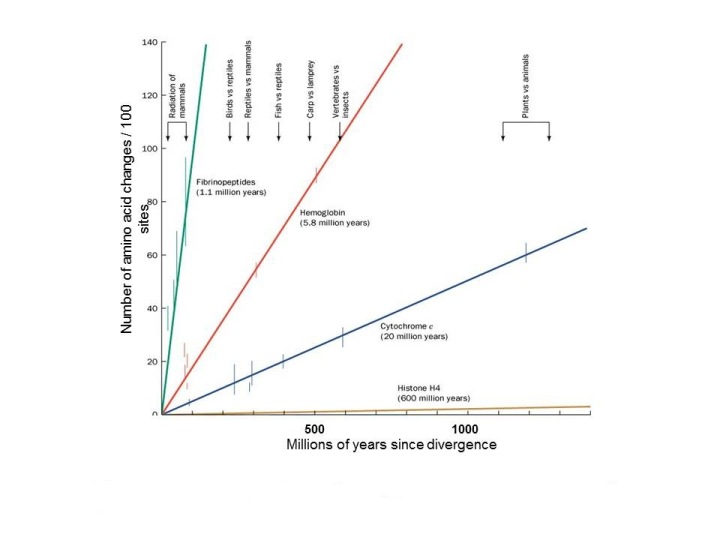
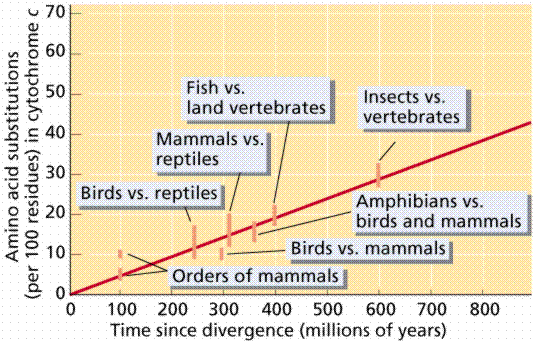
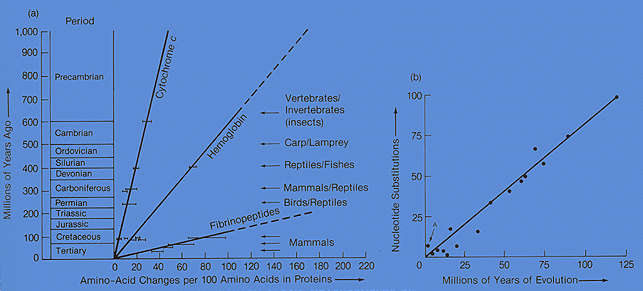

http://bio1100.nicerweb.com/Locked/media/ch14/molecular-cytochrome_c.html
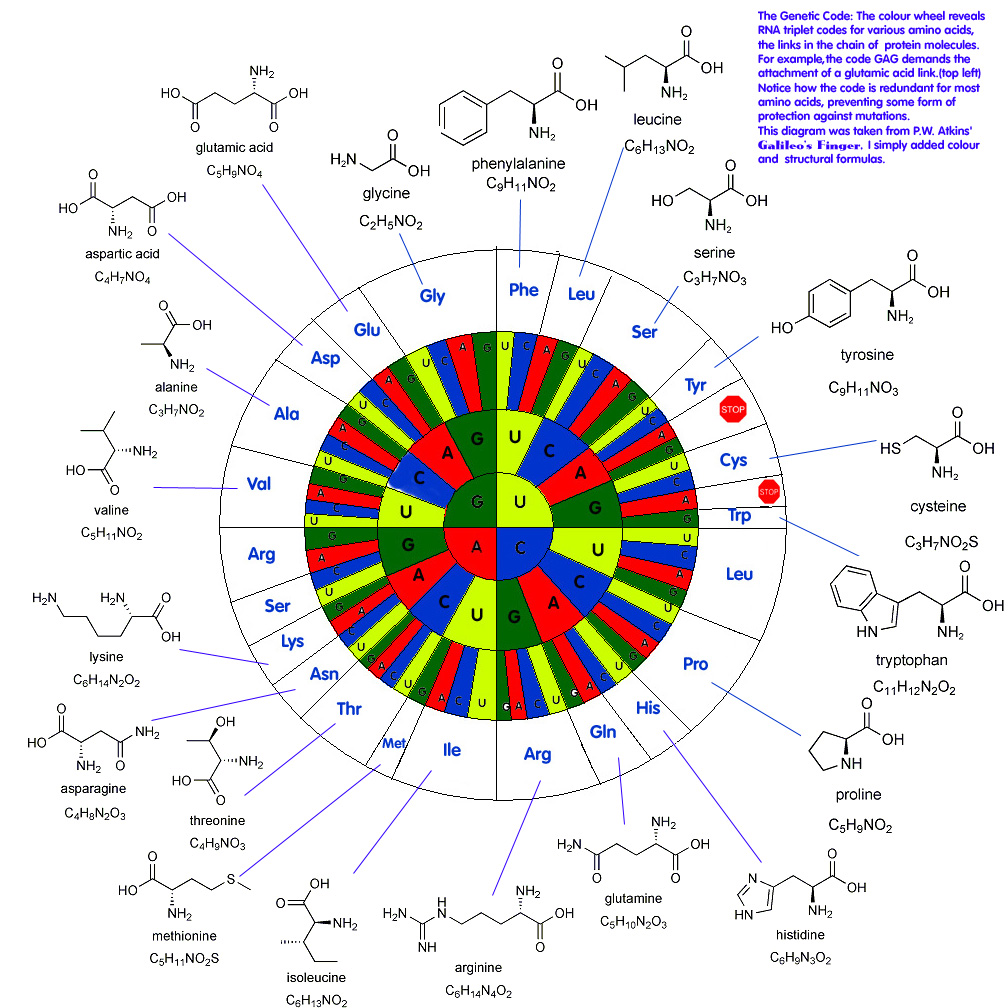
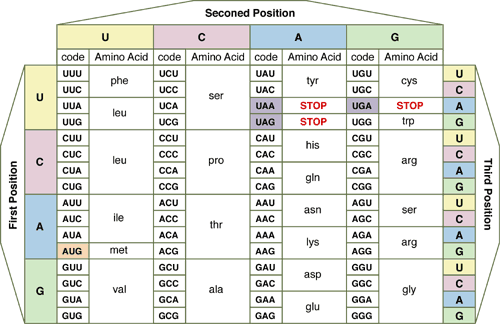
Genetic language similarities: Genes are transcribed from DNA into RNA and then are translated into amino acids which are put together to make protein. RNA used for translation comes in four “letters”– A, C, G, U (adenine, cytosine, guanine, and uracil). These letters are put together into three letter words called codons. The spelling of these words is consistent across almost all life (though it does not have to be at all!). This exchangeability makes genetic modification by recombinant DNA (DNA from one organism put into another) possible Here are several charts representing these words:
Biogeographic similarities: Species that are related to each other tend to live near each other. Why? Because their common ancestor was likely from a similar location.
For example, why are so many marsupial species just on Australia (the columns labeled ‘marsupial’ are from Australia and its vicinity)? Why are placental species on other continents? Why is their distribution so separate even though their appearance is so similar? The simplest and best explanation is that they evolved separately and convergently from different lineages.

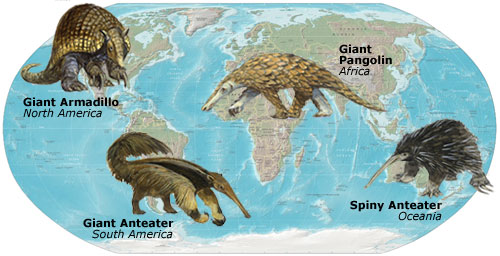
The same environmental niches often help create the same evolutionary adaptations, like these ant eating mammals.
This convergent evolution also results in similar species evolving separately (which we can verify with genetic testing) by analogous anole adaptations in the Caribbean and South America.

Likewise, in the cichlids of two different lakes: Lake Tanganyika (left) and Lake Malawi (right).

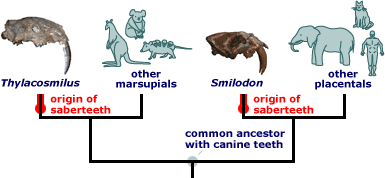
And in the saber toothed predators of Australia (Thylacosmilus–a marsupial) and North America (simlodon–a placental).